Referring to the Figure, What Bases Will Be Added to the Primer as Dna Replication Proceeds?
Affiliate ix: Introduction to Molecular Biological science
9.ii Deoxyribonucleic acid Replication
Learning Objectives
By the end of this section, you will be able to:
- Explain the procedure of Deoxyribonucleic acid replication
- Explain the importance of telomerase to DNA replication
- Draw mechanisms of Dna repair
When a cell divides, information technology is important that each daughter cell receives an identical copy of the DNA. This is accomplished by the procedure of Deoxyribonucleic acid replication. The replication of DNA occurs during the synthesis stage, or S phase, of the cell cycle, before the cell enters mitosis or meiosis.
The elucidation of the structure of the double helix provided a hint every bit to how DNA is copied. Recall that adenine nucleotides pair with thymine nucleotides, and cytosine with guanine. This ways that the two strands are complementary to each other. For instance, a strand of DNA with a nucleotide sequence of AGTCATGA will take a complementary strand with the sequence TCAGTACT (Figure 9.8).
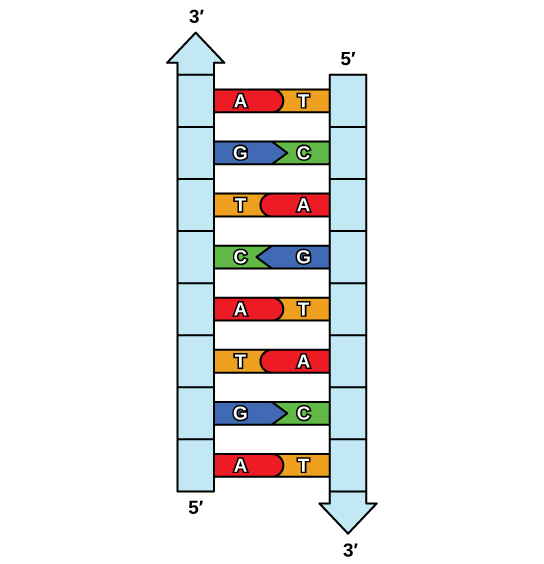
Because of the complementarity of the two strands, having one strand means that information technology is possible to recreate the other strand. This model for replication suggests that the two strands of the double helix carve up during replication, and each strand serves as a template from which the new complementary strand is copied (Effigy ix.9).
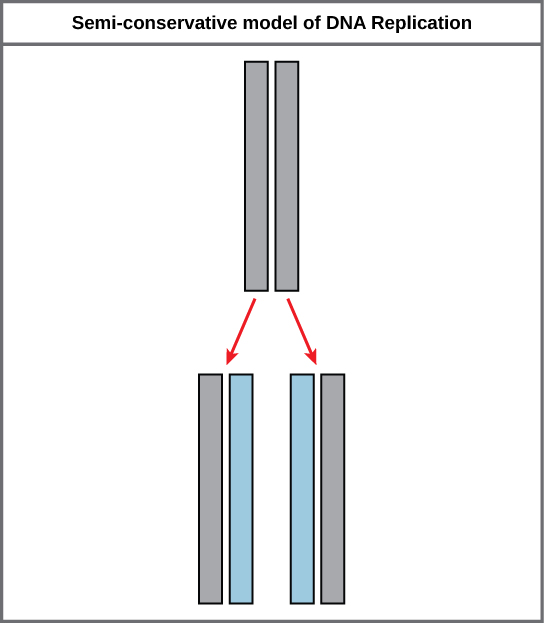
During DNA replication, each of the 2 strands that brand up the double helix serves as a template from which new strands are copied. The new strand volition be complementary to the parental or "old" strand. Each new double strand consists of one parental strand and ane new daughter strand. This is known equally semiconservative replication. When two DNA copies are formed, they have an identical sequence of nucleotide bases and are divided every bit into ii girl cells.
DNA Replication in Eukaryotes
Because eukaryotic genomes are very complex, Deoxyribonucleic acid replication is a very complicated process that involves several enzymes and other proteins. It occurs in 3 principal stages: initiation, elongation, and termination.
Call up that eukaryotic DNA is jump to proteins known as histones to form structures called nucleosomes. During initiation, the Deoxyribonucleic acid is made attainable to the proteins and enzymes involved in the replication procedure. How does the replication machinery know where on the Dna double helix to brainstorm? It turns out that in that location are specific nucleotide sequences called origins of replication at which replication begins. Certain proteins bind to the origin of replication while an enzyme called helicase unwinds and opens up the DNA helix. As the Dna opens up, Y-shaped structures called replication forks are formed (Figure ix.ten). Two replication forks are formed at the origin of replication, and these get extended in both directions equally replication proceeds. There are multiple origins of replication on the eukaryotic chromosome, such that replication can occur simultaneously from several places in the genome.
During elongation, an enzyme called Dna polymerase adds DNA nucleotides to the three′ end of the template. Because DNA polymerase tin only add new nucleotides at the end of a backbone, a primer sequence, which provides this starting point, is added with complementary RNA nucleotides. This primer is removed afterward, and the nucleotides are replaced with DNA nucleotides. Ane strand, which is complementary to the parental Dna strand, is synthesized continuously toward the replication fork so the polymerase tin add nucleotides in this direction. This continuously synthesized strand is known as the leading strand. Because DNA polymerase can only synthesize Dna in a v′ to 3′ direction, the other new strand is put together in brusque pieces called Okazaki fragments. The Okazaki fragments each crave a primer made of RNA to get-go the synthesis. The strand with the Okazaki fragments is known as the lagging strand. As synthesis proceeds, an enzyme removes the RNA primer, which is and then replaced with DNA nucleotides, and the gaps between fragments are sealed by an enzyme called Deoxyribonucleic acid ligase.
The process of Deoxyribonucleic acid replication can be summarized as follows:
- Dna unwinds at the origin of replication.
- New bases are added to the complementary parental strands. One new strand is made continuously, while the other strand is made in pieces.
- Primers are removed, new Dna nucleotides are put in place of the primers and the backbone is sealed by DNA ligase.
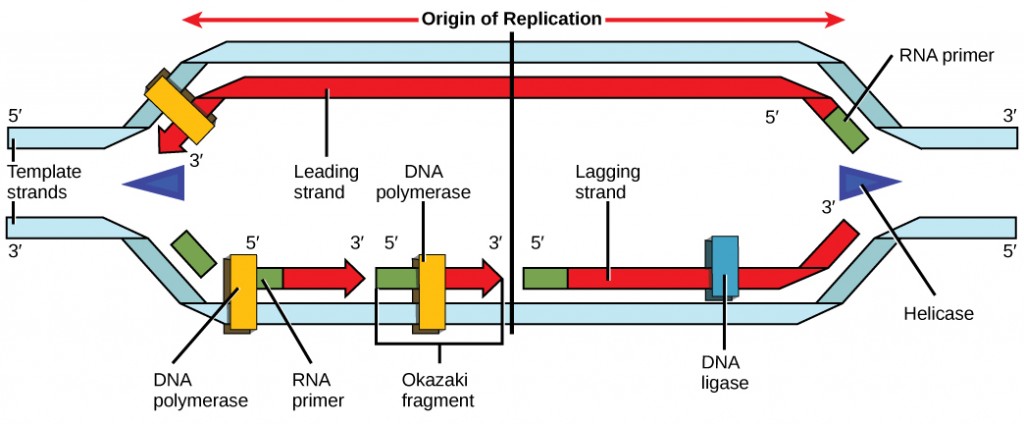
You isolate a cell strain in which the joining together of Okazaki fragments is dumb and suspect that a mutation has occurred in an enzyme found at the replication fork. Which enzyme is virtually likely to be mutated?
Telomere Replication
Considering eukaryotic chromosomes are linear, Deoxyribonucleic acid replication comes to the end of a line in eukaryotic chromosomes. As y'all have learned, the Dna polymerase enzyme can add nucleotides in only one management. In the leading strand, synthesis continues until the end of the chromosome is reached; however, on the lagging strand there is no place for a primer to exist fabricated for the DNA fragment to be copied at the end of the chromosome. This presents a problem for the cell because the ends remain unpaired, and over fourth dimension these ends get progressively shorter as cells proceed to carve up. The ends of the linear chromosomes are known as telomeres, which have repetitive sequences that do not code for a detail gene. Equally a result, it is telomeres that are shortened with each round of DNA replication instead of genes. For example, in humans, a half dozen base-pair sequence, TTAGGG, is repeated 100 to 1000 times. The discovery of the enzyme telomerase (Figure 9.11) helped in the understanding of how chromosome ends are maintained. The telomerase attaches to the end of the chromosome, and complementary bases to the RNA template are added on the stop of the Deoxyribonucleic acid strand. Once the lagging strand template is sufficiently elongated, DNA polymerase can at present add nucleotides that are complementary to the ends of the chromosomes. Thus, the ends of the chromosomes are replicated.
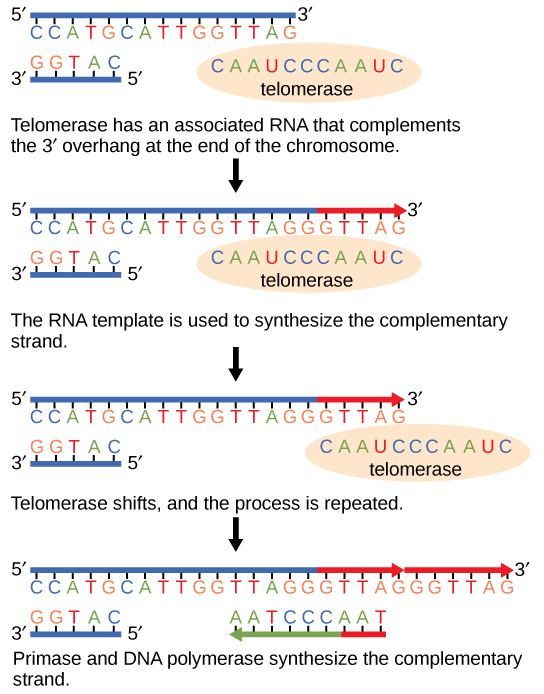
Telomerase is typically found to exist active in germ cells, developed stalk cells, and some cancer cells. For her discovery of telomerase and its action, Elizabeth Blackburn (Figure nine.12) received the Nobel Prize for Medicine and Physiology in 2009.

Telomerase is not active in adult somatic cells. Adult somatic cells that undergo prison cell partitioning continue to have their telomeres shortened. This essentially means that telomere shortening is associated with aging. In 2010, scientists found that telomerase can reverse some age-related atmospheric condition in mice, and this may have potential in regenerative medicine. one Telomerase-deficient mice were used in these studies; these mice have tissue atrophy, stalk-cell depletion, organ arrangement failure, and dumb tissue injury responses. Telomerase reactivation in these mice caused extension of telomeres, reduced Deoxyribonucleic acid damage, reversed neurodegeneration, and improved functioning of the testes, spleen, and intestines. Thus, telomere reactivation may have potential for treating age-related diseases in humans.
DNA Replication in Prokaryotes
Recall that the prokaryotic chromosome is a circular molecule with a less extensive coiling structure than eukaryotic chromosomes. The eukaryotic chromosome is linear and highly coiled around proteins. While at that place are many similarities in the DNA replication process, these structural differences necessitate some differences in the DNA replication process in these two life forms.
DNA replication has been extremely well-studied in prokaryotes, primarily because of the small size of the genome and large number of variants available. Escherichia coli has 4.six million base of operations pairs in a single circular chromosome, and all of it gets replicated in approximately 42 minutes, starting from a single origin of replication and proceeding around the chromosome in both directions. This means that approximately thou nucleotides are added per second. The process is much more rapid than in eukaryotes. The table below summarizes the differences between prokaryotic and eukaryotic replications.
| Holding | Prokaryotes | Eukaryotes |
|---|---|---|
| Origin of replication | Unmarried | Multiple |
| Rate of replication | grand nucleotides/s | 50 to 100 nucleotides/due south |
| Chromosome structure | circular | linear |
| Telomerase | Non present | Present |
Concept in Action

Click through a tutorial on Deoxyribonucleic acid replication.
Deoxyribonucleic acid Repair
Dna polymerase can make mistakes while calculation nucleotides. Information technology edits the Dna by proofreading every newly added base of operations. Wrong bases are removed and replaced past the correct base, and so polymerization continues (Figure 9.13 a). Almost mistakes are corrected during replication, although when this does not happen, the mismatch repair mechanism is employed. Mismatch repair enzymes recognize the wrongly incorporated base of operations and excise it from the Dna, replacing information technology with the correct base (Effigy nine.xiii b). In nevertheless another type of repair, nucleotide excision repair, the DNA double strand is unwound and separated, the incorrect bases are removed along with a few bases on the 5′ and 3′ end, and these are replaced by copying the template with the help of Deoxyribonucleic acid polymerase (Figure 9.13 c). Nucleotide excision repair is peculiarly important in correcting thymine dimers, which are primarily caused by ultraviolet light. In a thymine dimer, two thymine nucleotides adjacent to each other on 1 strand are covalently bonded to each other rather than their complementary bases. If the dimer is not removed and repaired information technology will pb to a mutation. Individuals with flaws in their nucleotide excision repair genes bear witness extreme sensitivity to sunlight and develop skin cancers early in life.
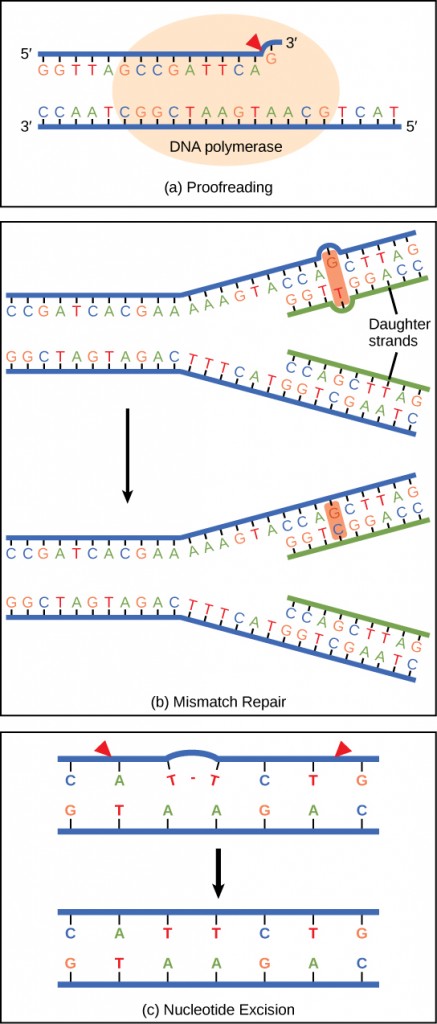
Nearly mistakes are corrected; if they are not, they may result in a mutation—defined as a permanent change in the Dna sequence. Mutations in repair genes may lead to serious consequences like cancer.
Section Summary
DNA replicates by a semi-conservative method in which each of the two parental Dna strands act as a template for new Dna to exist synthesized. After replication, each DNA has i parental or "old" strand, and one girl or "new" strand.
Replication in eukaryotes starts at multiple origins of replication, while replication in prokaryotes starts from a single origin of replication. The Deoxyribonucleic acid is opened with enzymes, resulting in the formation of the replication fork. Primase synthesizes an RNA primer to initiate synthesis by Dna polymerase, which tin add nucleotides in only i management. One strand is synthesized continuously in the direction of the replication fork; this is chosen the leading strand. The other strand is synthesized in a management abroad from the replication fork, in short stretches of Deoxyribonucleic acid known as Okazaki fragments. This strand is known as the lagging strand. One time replication is completed, the RNA primers are replaced by DNA nucleotides and the DNA is sealed with Deoxyribonucleic acid ligase.
The ends of eukaryotic chromosomes pose a problem, as polymerase is unable to extend them without a primer. Telomerase, an enzyme with an inbuilt RNA template, extends the ends by copying the RNA template and extending ane end of the chromosome. DNA polymerase can then extend the Deoxyribonucleic acid using the primer. In this way, the ends of the chromosomes are protected. Cells have mechanisms for repairing DNA when information technology becomes damaged or errors are made in replication. These mechanisms include mismatch repair to supervene upon nucleotides that are paired with a non-complementary base and nucleotide excision repair, which removes bases that are damaged such equally thymine dimers.
Glossary
DNA ligase: the enzyme that catalyzes the joining of DNA fragments together
DNA polymerase: an enzyme that synthesizes a new strand of Deoxyribonucleic acid complementary to a template strand
helicase: an enzyme that helps to open up the Dna helix during DNA replication by breaking the hydrogen bonds
lagging strand: during replication of the 3′ to five′ strand, the strand that is replicated in short fragments and away from the replication fork
leading strand: the strand that is synthesized continuously in the 5′ to 3′ direction that is synthesized in the direction of the replication fork
mismatch repair: a form of DNA repair in which non-complementary nucleotides are recognized, excised, and replaced with right nucleotides
mutation: a permanent variation in the nucleotide sequence of a genome
nucleotide excision repair: a form of Dna repair in which the DNA molecule is unwound and separated in the region of the nucleotide damage, the damaged nucleotides are removed and replaced with new nucleotides using the complementary strand, and the DNA strand is resealed and allowed to rejoin its complement
Okazaki fragments: the DNA fragments that are synthesized in short stretches on the lagging strand
primer: a short stretch of RNA nucleotides that is required to initiate replication and allow Dna polymerase to bind and begin replication
replication fork: the Y-shaped structure formed during the initiation of replication
semiconservative replication: the method used to replicate Dna in which the double-stranded molecule is separated and each strand acts as a template for a new strand to be synthesized, and then the resulting Deoxyribonucleic acid molecules are composed of ane new strand of nucleotides and 1 old strand of nucleotides
telomerase: an enzyme that contains a catalytic office and an inbuilt RNA template; it functions to maintain telomeres at chromosome ends
telomere: the DNA at the end of linear chromosomes
Footnotes
ane Mariella Jaskelioff, et al., "Telomerase reactivation reverses tissue degeneration in aged telomerase-deficient mice," Nature, 469 (2011):102–seven.
Source: https://opentextbc.ca/biology/chapter/9-2-dna-replication/
0 Response to "Referring to the Figure, What Bases Will Be Added to the Primer as Dna Replication Proceeds?"
Post a Comment